Projects
-
Revealing Hidden Insights: Deep Learning Recovery of SWI Data to Explore Brain Iron Levels and Neurodevelopment in Preterm Infants
Funding: NSERC Discovery Grant
Assignees: Floria Lu
Background: When babies are born very preterm (2-4 months early), the brain development that typically occurs in the womb takes place instead in the neonatal intensive care unit (NICU). During this period, brain injuries and exposure to repetitive pain and stress can have lasting effects on their neurodevelopment, which researchers are only beginning to understand.1 Advances in medical imaging have provided invaluable tools for studying the developing brain, yet these tools often fall short of their full potential. For instance, Susceptibility Weighted Imaging (SWI) – a magnetic resonance imaging (MRI) sequence commonly used to clinically detect subtle brain injuries – inherently discards valuable quantitative information.2 As a result, these scans are qualitative in nature, meaning pixel values cannot be directly compared across individuals, or used for precise measurements. This discarded information, however, holds significant promise. If we could preserve and analyze the unfiltered SWI data, researchers could apply a method known as Quantitative Susceptibility Mapping (QSM) to extract quantitative measures of venous oxygenation, myelination, iron content, and more. These metrics are critical for answering pressing scientific questions about brain development and injury.3 For example, iron needs to be provided in adequate amounts – not too much and not too little – due to its importance for a remarkable array of essential functions during this period.4 Recent advances in artificial intelligence (AI), particularly deep learning (DL), are unlocking new possibilities for recovering this lost information. While large language models such as ChatGPT have captured public attention, DL algorithms are making equally groundbreaking contributions to clinical research. Specifically, DL-based "unfiltering" techniques can restore the data lost during the creation of clinical SWI scans, enabling researchers to perform QSM analyses retrospectively on existing datasets.5,6 To explore this potential, we propose to test several published DL unfiltering algorithms on data acquired through the Preterm Care Study, a collaborative effort led by Drs. Steven Miller and Ruth Grunau. These researchers, and many others around the world, currently lack the tools to unlock the full potential of their SWI data, but with DL-based approaches, this limitation could be overcome, paving the way for new insights into preterm brain development and care. Hypothesis We hypothesize that reduced iron levels in the basal ganglia and deep gray matter, measured using QSM calculated from DL recovered unfiltered SWI data, will correlate with reduced motor and cognitive scores on the Bayley Scales of Infant and Toddler Development in the same infants at 18 months. Aims and Methodology Aims: Apply DL algorithms on the filtered SWI data to recover the unfiltered phase data; Use the unfiltered SWI data to create QSM images in order to investigate correlations of iron levels in deep grey matter of very preterm infants with neurodevelopmental outcomes later in life.
Methods: Approximately 120 infants born <32 weeks' gestational age were recruited at BC Women’s Hospital NICU between 2015 and 2019. Imaging took place at BCCH Research MRI Facility on their 3T scanner. Some infants were scanned shortly after birth, some at term-equivalent age, and approximately 60 were scanned at both time-periods. 3D SWI sequences (0.625x0.625x1 mm3) were originally acquired in order to detect, locate, and measure the size of injuries such as brain bleeds and white matter injuries. As well, 3D anatomical scans were acquired, which will be used to segment the brain and isolate the deep grey matter structures using state-of-the-art neonatal segmentation pipelines.7 We plan to make use of previously published DL algorithms5,6, as well as weights that have already been trained on our scanner (github.com/kamesy/homodynenet). QSM images will then be calculated using the recovered unfiltered-phase and the superior rapid two-step dipole inversion method8 and open-source software (github.com/kamesy/QSM.m). Using the deep grey-matter segmentation masks, average magnetic susceptibility values will be calculated for all scans.
Statistics: The statistical software R will be used to assess whether a relationship exists between motor and cognitive scores at 18 months and deep gray matter iron levels at preterm and term-equivalent ages. A mixed-effects multiple linear regression model will be employed, treating subjects as random effects to account for the longitudinal nature of the data. Gestational age, sex, number of painful procedures while in the NICU, total brain volume, maternal age at birth, parental socioeconomic status, parental educational level will be also included as covariates to control for and explore potential confounding factors. Significance By testing and validating these algorithms on data from the Preterm Care Study, this project represents an opportunity to harness cutting-edge computational techniques to address pressing questions in preterm brain development. Applying these DL-based unfiltering algorithms to existing datasets with SWI scans would allow researchers to quantify valuable information such as venous oxygenation, myelination, and iron levels retrospectively in hundreds of preterm and neonatal studies worldwide. Feasibility All of the data has already been collected and is available to download from BCCHR’s XNAT server, meaning the most difficult part of the study has already concluded. If the pre-trained DL weights do not produce satisfactory images (i.e. result in noisy QSM images), we have access to unfiltered SWI data in neonates that we can use to train our own weights (data previously collected by Drs. Weber and Grunau; n = 19 infants born very preterm and scanned at term-equivalent age at the same hospital and research MRI facility). -
Investigating Brain Dynamics and Power Law Scaling in fMRI BOLD Signals
Funding: NSERC Discovery Grant; BCCH Foundation
Assignees: Erhan Asad Javed
Background: Since the 1990s, it has been suggested that the brain operates in a critical state between order and disorder. Systems that exist in a critical state exhibit scale-invariance, power-law distributions, fractal geometry, and long-range correlations. Evidence to support the brain criticality hypothesis comes from many different avenues, both spatial (dendritic branching and small-world network connectivity), and temporal (neurotransmitter release, neuronal firing, local field potentials, and MEG, EEG, and fMRI signals), suggesting that in the brain, criticality is the rule, not the exception. It has been further theorized that existing in this state maximizes signalling efficiency, storage capacity, and flexibility. Thus, being able to properly identify and measure criticality is a key component in trying to understand how the brain functions. Unfortunately, in a rush to investigate and publish findings on criticality in fMRI neuroimaging, a rigorous testing of the assumptions and algorithms used for these purposes has not yet been performed.
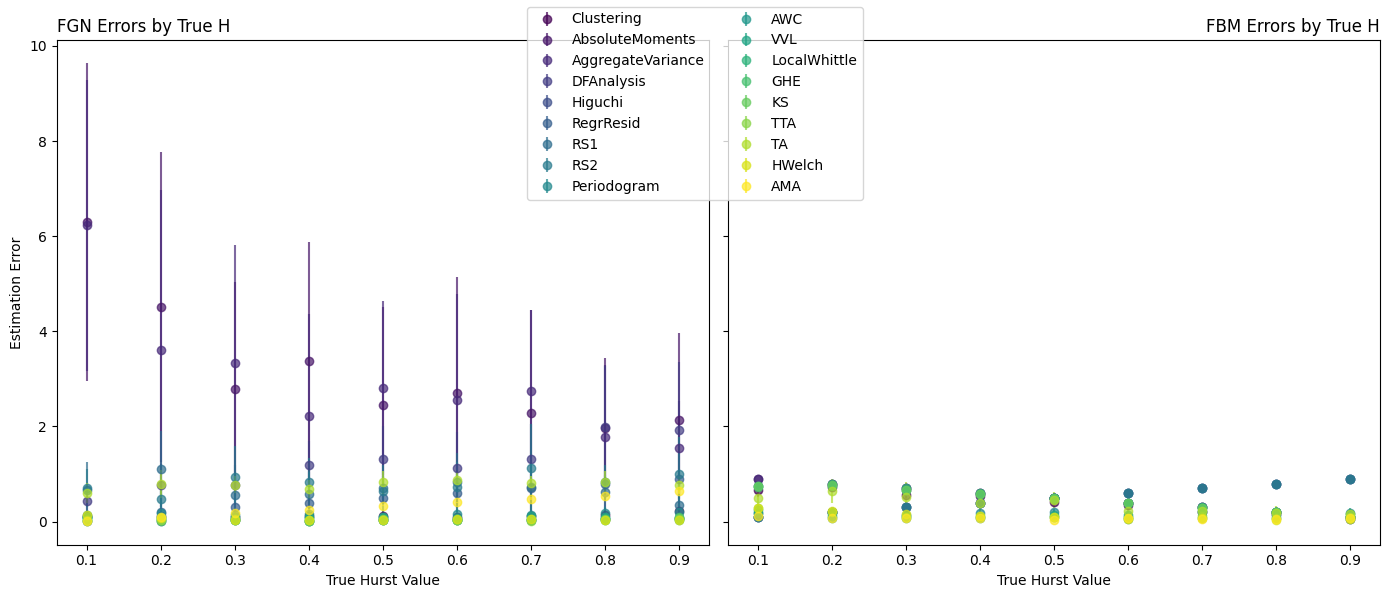
Objective: The objective of this student project is to develop a signal-processing tool in Python that will: 1) test whether a signal follows a power-law distribution; 2) categorize the signal as either fractional Gaussian noise (fGn) or fractional Brownian motion (fBm); and 3) accurately report the signal’s Hurst exponent (H) value between 0 and 1. Time permitting, the tool will be used to investigate the nature of fMRI signals and the effects of various preprocessing methods.
Methods: Where possible, previously published Python libraries will be used to simulate power-law distributed signals of known type (fGn vs fBm) and H values. An exhaustive search for previously published H estimation algorithms will be conducted and converted to Python where necessary. Testing will be performed throughout the development process to ensure reliability and performance.
Deliverables: The following deliverables will be provided at the end of the project: 1) A signal pro- cessing tool that includes all of the features described above as well as: can analyze a 4D fMRI file with ∼250,000 voxels in approximately 10 minutes; provides easy to read visual outputs for verification and publication; is flexible and easy to use; has code-base that is well formatted and with easy to understand comments throughout; and is version controlled on a public Github repository; 2) Documentation for the tool, including installation instructions and user instructions; 3) Test cases and testing documentation; 4) A manuscript draft to be submitted for peer-review and published for wider knowledge-translation and dissemination. -
Functional, Metabolic, and Structural MRI Findings in Rett Syndrome
Funding: British Columbia Children’s Hospital Research Institute (Establishment Award and Salaries), Ontario Rett Syndrome Association Research Grant
Collaborators: Dr. Anita Datta, Dr. Gabriella Horvath
Background: Rett Syndrome (RTT) is a severe genetic neurodevelopmental disorder that typically manifests in females due to de novo mutations in the methyl-CpG-binding protein 2 gene (MECP2). Symptoms usually become apparent after 6 to 18 months of age when the child begins to miss developmental milestones or loses abilities they had previously gained. These symptoms typically affect nearly every aspect of the child’s life. There is no cure for Rett syndrome, treatment is symptomatic only. In order to design better treatment, further research is necessary to understand anatomical, neurochemical, and functional changes in the brains of children with RTT, and to link these changes with measures of genetics, disease progression and clinical features.
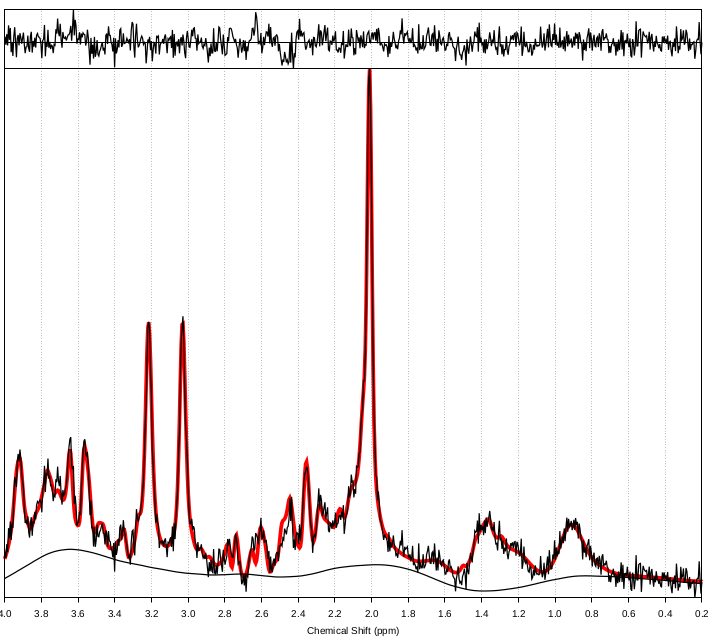
Aim: The primary aim of the current study will be to investigate in vivo white matter architectural (DTI) and metabolism (MRS) differences between girls with RTT compared to healthy age-matched girls, as well as explore the relationship between these neuroimaging measures and clinical severity scores, epilepsy severity, and genetic MECP2 mutations. Secondary aims will explore functional MRI differences across RTT severity and genetic mutations. This data will provide critical preliminary data for a full Project Grant application to CIHR.
Methods: Girls with RTT (aged 8-12 years; n = 10) will be recruited from BC Children’s Hospital (Dr. Datta and Dr. Horvath’s clinics), as well as from the Provincial Rett Clinic. Healthy controls (aged 8-12 years; n = 10) will be recruited from social media, and from friends of families with RTT. MRI will be acquired on the BC Children’s Hospital MRI Research Facility research dedicated broadband GE 3T MRI system. EEG and biochemical data will be collected as part of clinical care.
BB&D Theme: Our goals are aligned with the vision of BB&D to improve outcomes for developmental and mental health of at risk children. This catalyst grant will help researchers Dr. Weber and Datta to collaborate on a project that will gather and test new analyses of MRI and clinical/genetic data of children with complex medical, developmental and mental health needs. This research is in child neurodevelopmental health.
Expected Outcomes: This research will help identify the structural, metabolic and functional differences between girls with RTT and their healthy counterparts. These findings will be linked to specific MECP2 mutations and clinical severity, allowing for a greater understanding of the specific causes of RTT, and paving the way for future targeted therapies. The overarching goal will be to scale this project up after we have collected the pilot data and shown proof of concept.